Publications
Group highlights
(For a larger list of publications see below or go to Google Scholar.
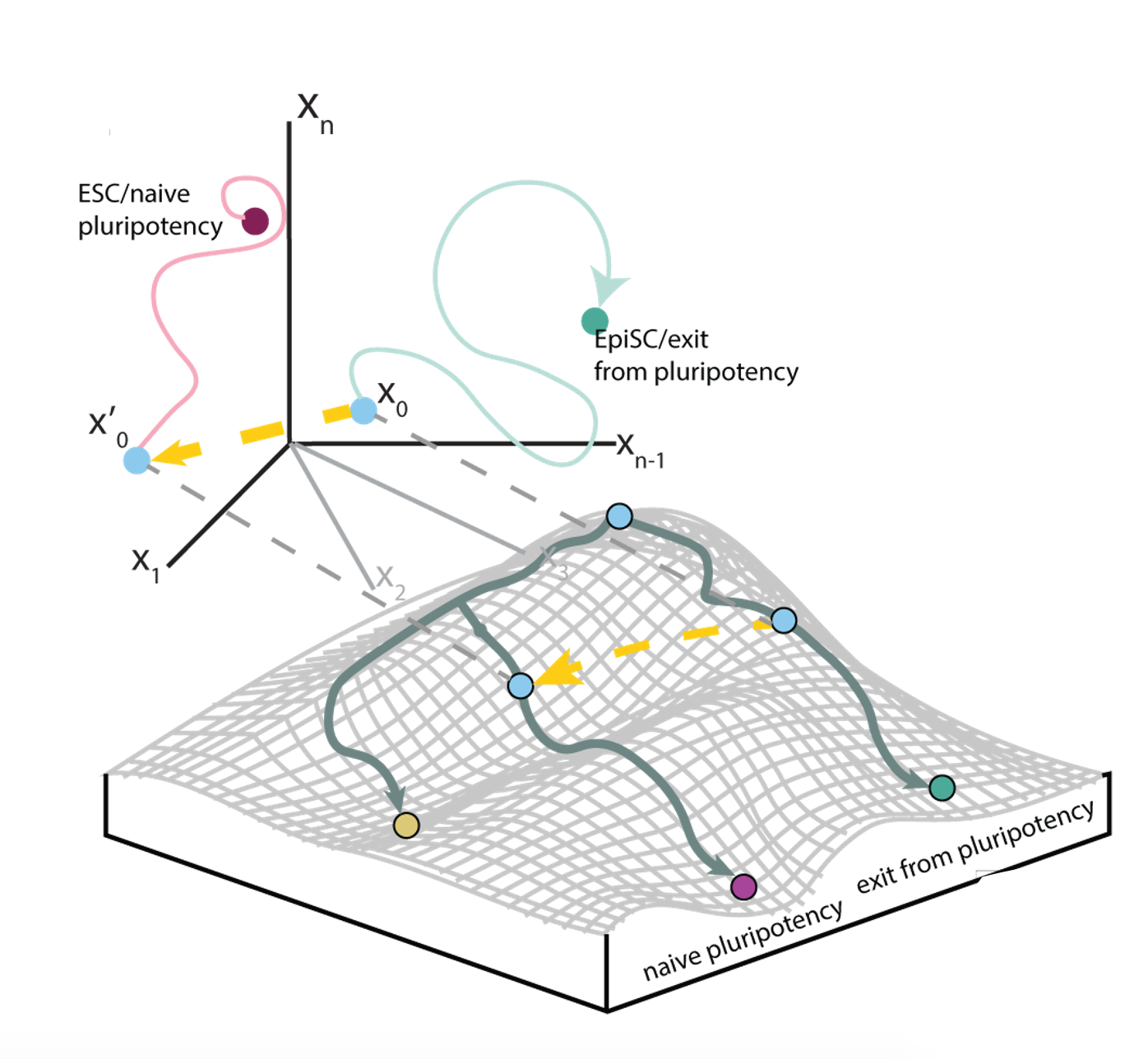
We present NETISCE, a novel computational tool for identifying cell fate reprogramming targets in static networks. In combination with machine learning algorithms, NETISCE estimates the attractor landscape and predicts reprogramming targets using Signal Flow Analysis and Feedback Vertex Set Control, respectively.
Lauren Marazzi, Milan Shah, Shreedula Balakrishnan, Ananya Patil, Paola Vera-Licona
npj Systems Biology and Applications volume 8, Article number 21 (2022)
NETISCE software tool can be found here.
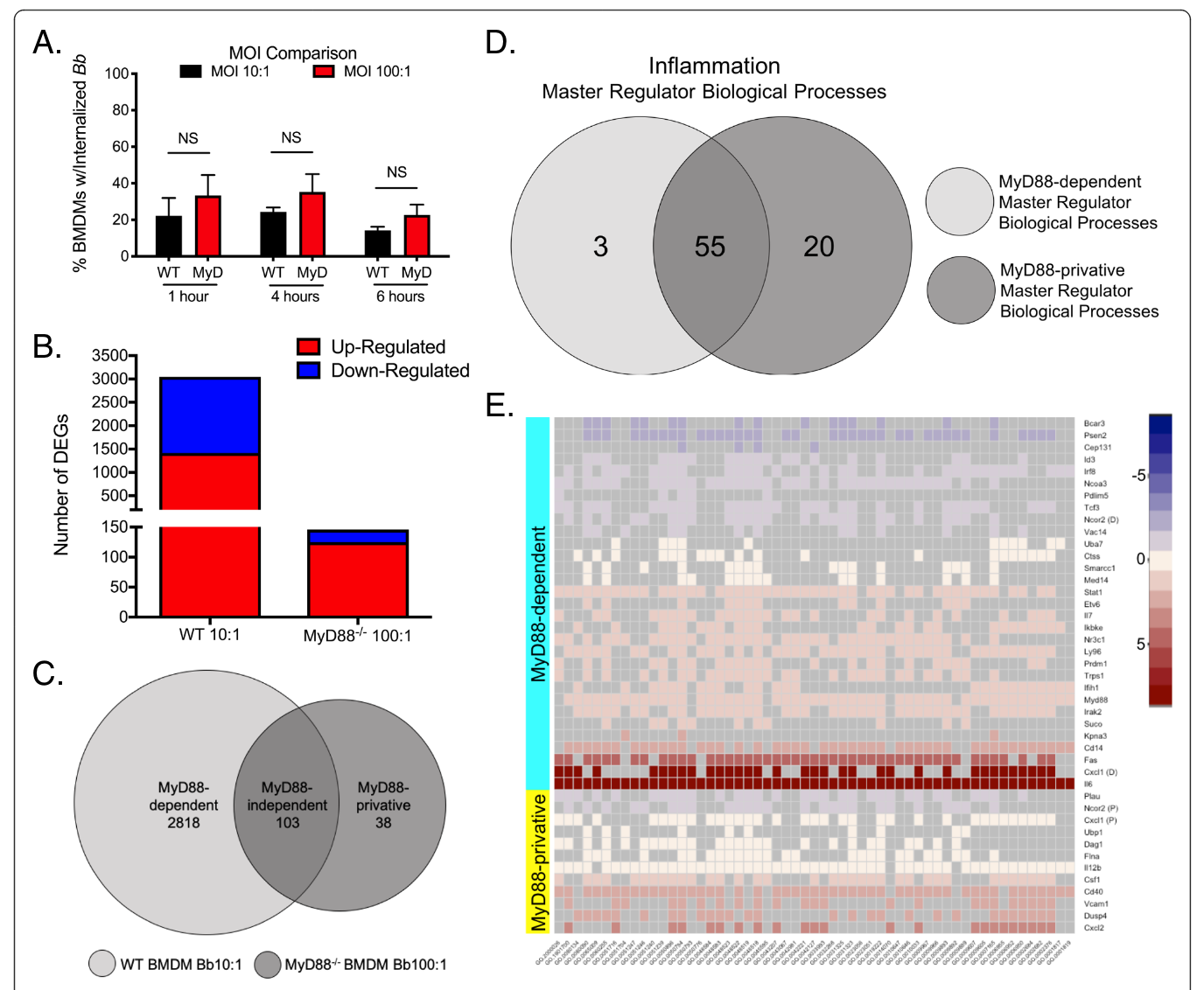
First paper from our collaboration with the Salazar Lab. We were in charge of the data-driven reconstruction and analysis of the signaling networks.
Sarah J. Benjamin, Kelly L. Hawley, Paola Vera-Licona, Carson J. La Vake, Jorge L. Cervantes, Yijun Ruan, Justin D. Radolf and Juan C. Salazar
BMC Immunology volume 22, Article number 32 (2021)
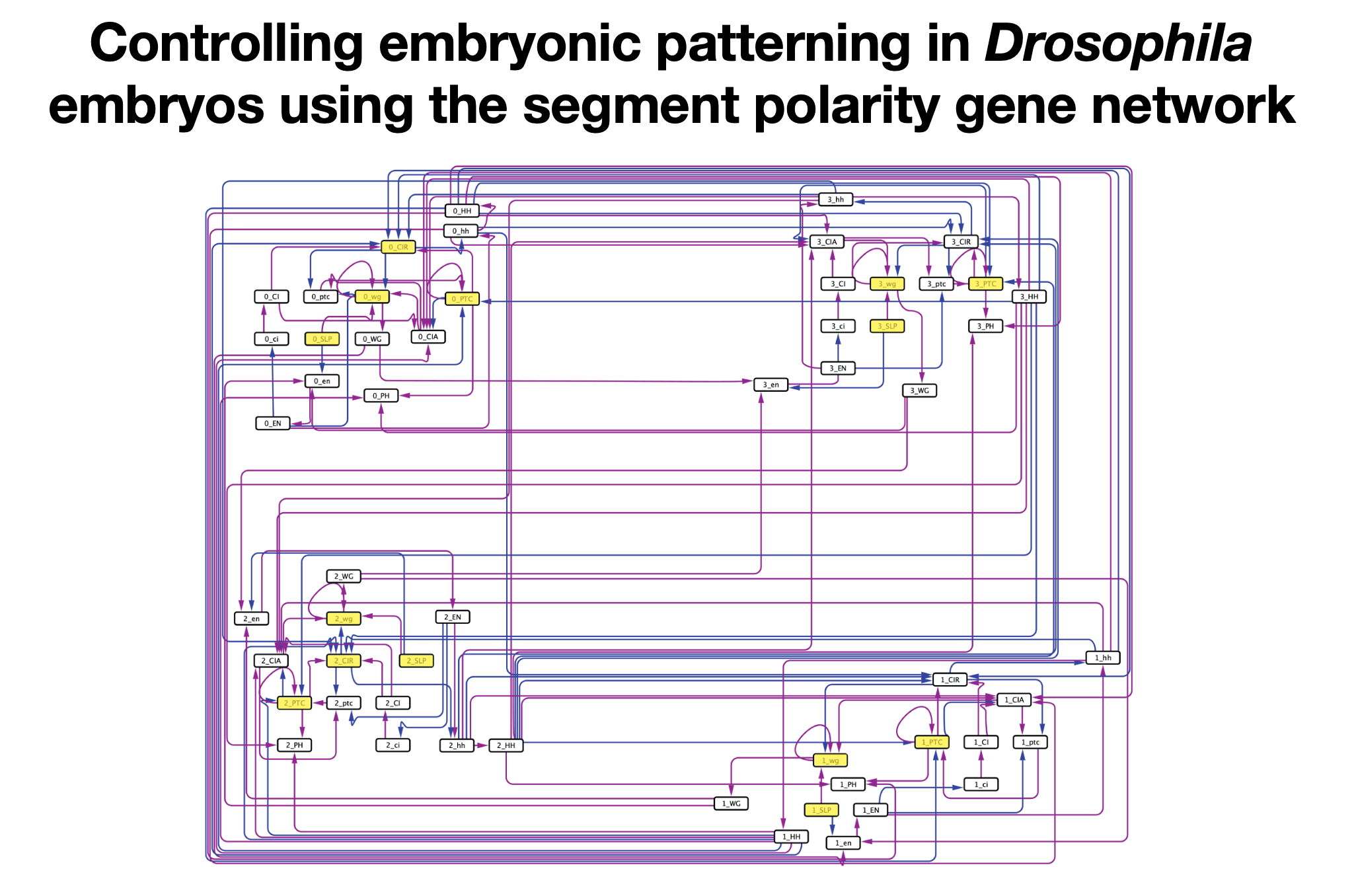
OCSANA+ is a Cytoscape app for identifying nodes to drive the system toward a desired long-term behavior, prioritizing combinations of interventions in large-scale complex networks, and estimating the effects of node perturbations in signaling networks, all based on the analysis of the network’s structure.
Lauren Marazzi, Andrew Gainer-Dewar, Paola Vera-Licona
Bioinformatics, Volume 36, Issue 19 (2020)
OCSANA+ software tool can be found here.
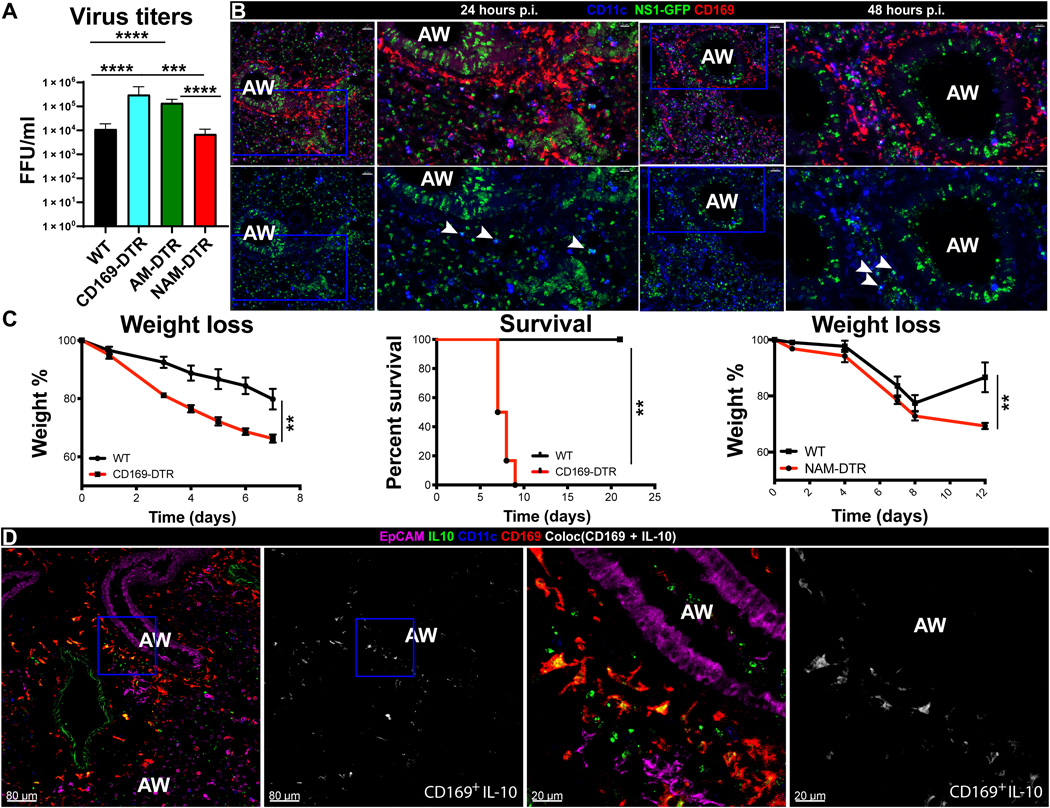
Our second paper from our collaboration with the Khanna Lab. We were in charge of the RNA-sequencing data analysis.
Basak B Ural,Stephen T Yeung, Payal Damani-Yokota, Joseph C Devlin, Maren de Vries, Paola Vera-Licona, Tasleem Samji, Catherine M Sawai, Geunhyo Jang, Oriana A Perez, Quynh Pham, Leigh Maher Png Loke, Meike Dittmann, Boris Reizis and Kamal M. Khanna
Science Immunology, Vol 5, Issue 45 (2020)
News coverage by Scientific American, AAAS, Technology Networks, Inverse, Pour La Science, and La Recherche. Selected in the Top 10 Health Innovations of 2020 by Health Innovations
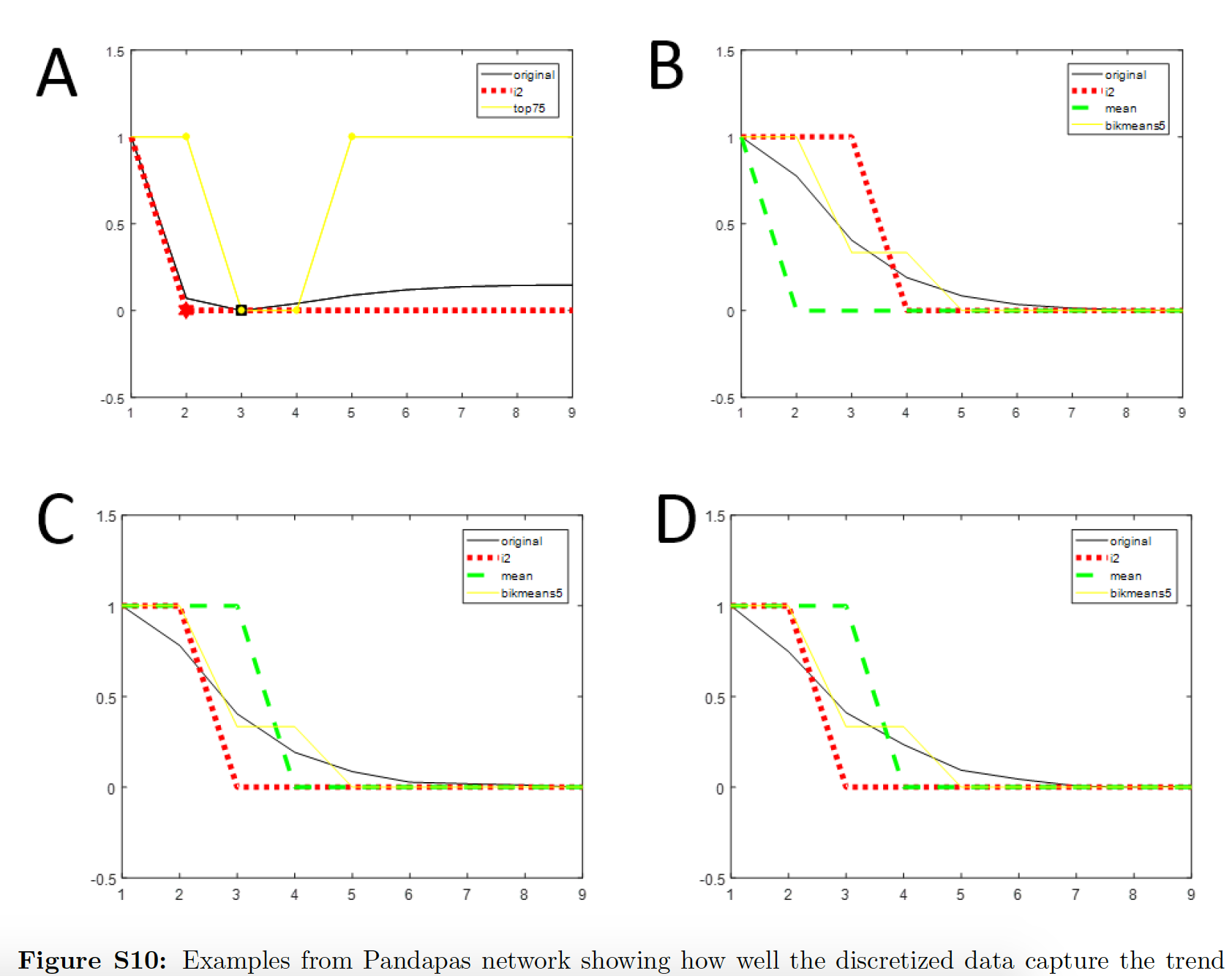
In this paper we introduce DiscreeTest, a two-step evaluation metric for ranking discretization methods for time-series data. To our knowledge, this is the first time that a method for benchmarking and selecting an appropriate discretization method for time-series data has been proposed.
Yuezhe Li, Tiffany Jann, Paola Vera-Licona
Bioinformatics, Volume 35, Issue 17 (2019)
DiscreeTest code can be found here.
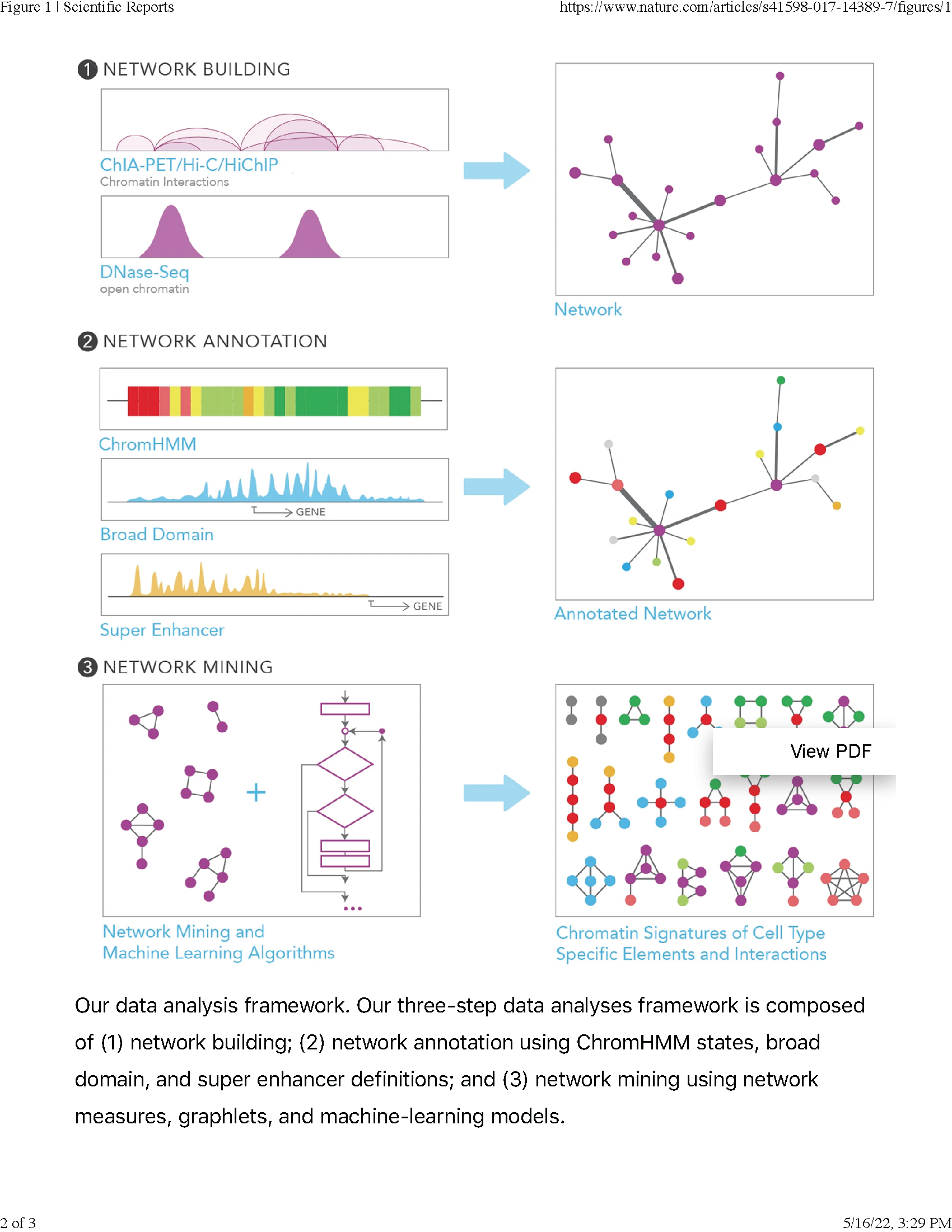
In this paper from our collaboration with the Ucar Lab, we studied chromatin interaction networks. We contributed to this paper by applying the less commonly used network theory concepts of graphlets and their orbits to describe and characterize the connectivity patterns of nodes in chromatin interaction networks.
Asa Thibodeau, Eladio J Márquez, Dong-Guk Shin, Paola Vera-Licona, Duygu Ucar
Scientific Reports volume 7, Article number 14466 (2017)
This paper was Selected Editors Choice, Epigenetics
Our first paper from our collaboration with the Khanna Lab. We were in charge of the RNA-sequencing data analysis.
Oriana Perez, Stephen Yeung, Paola Vera-Licona, Pablo Romagnoli, Tasleem Samji, Basak B Ural, Leigh Maher, Masato Tanaka, Kamal M Khanna
Science Immunology, Vol 2, Issue 16 (2017)
News coverage by Scientific American and Pour la Science . Article featured on the magazine cover

Finding inclusion-minimal hitting sets (MHSs) for a given family of sets is a fundamental combinatorial problem with applications in domains as diverse as Boolean algebra, computational biology, and data mining. In this paper we survey and benchmark several algorithms for MHS generation from across a variety of domains. We find that the fastest algorithms in practice are not those with the tightest complexity bounds or those most commonly used in applications.
Andrew Gainer-Dewar, Paola Vera-Licona
SIAM Journal on Discrete Mathematics Vol. 31 (2017)
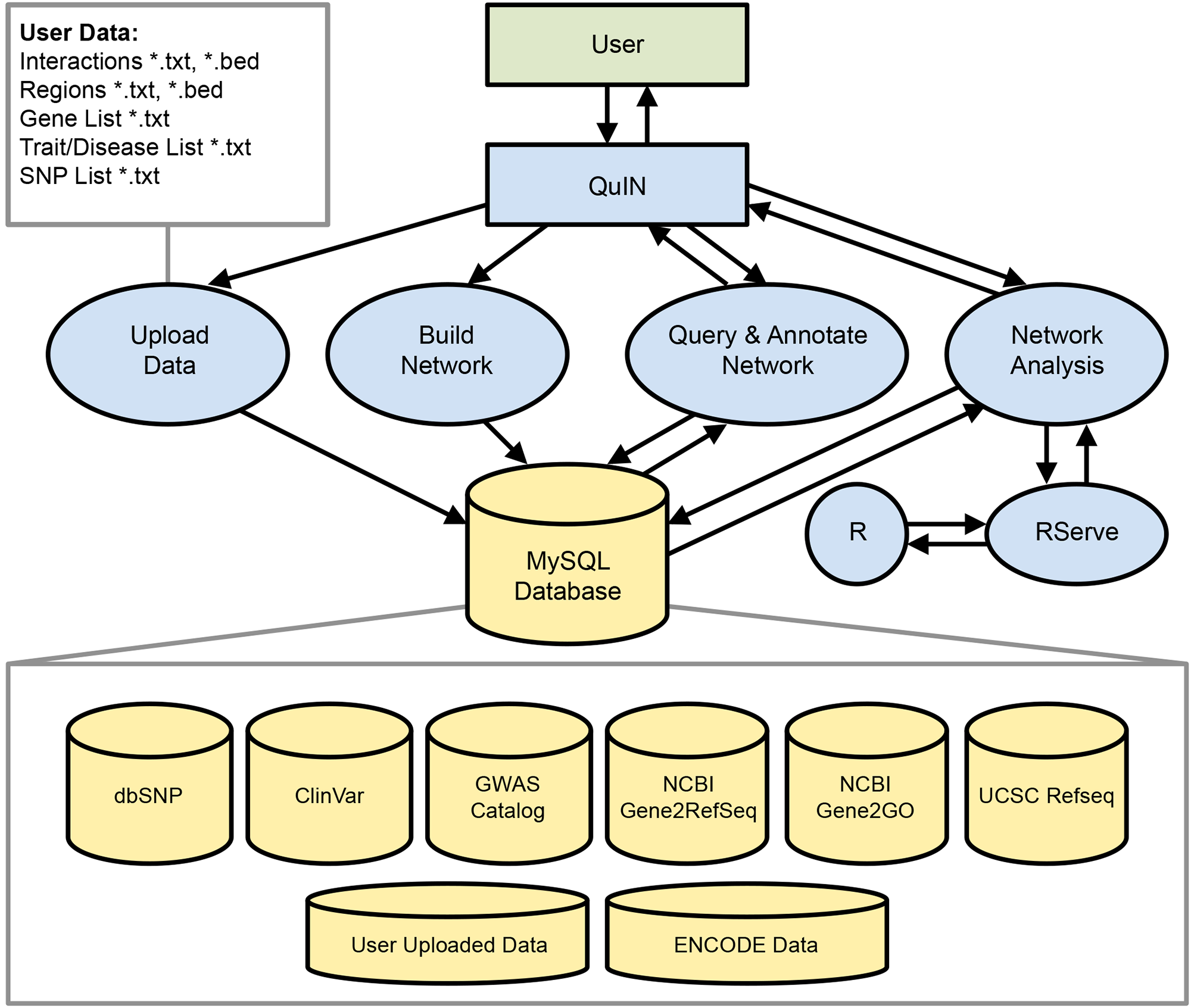
First paper from our collaboration with the Ucar Lab to develop a tool for querying and visualizing chromatin interaction networks. This is the first publicly available software for analysis of chromatin interaction networks that is web-accessible and easy to use, making it suitable for molecular biologists with no programming experience.
Asa Thibodeau, Eladio J Márquez, Oscar Luo, Yijun Ruan, Francesca Menghi, Dong-Guk Shin, Michael L Stitzel, Paola Vera-Licona, Duygu Ucar
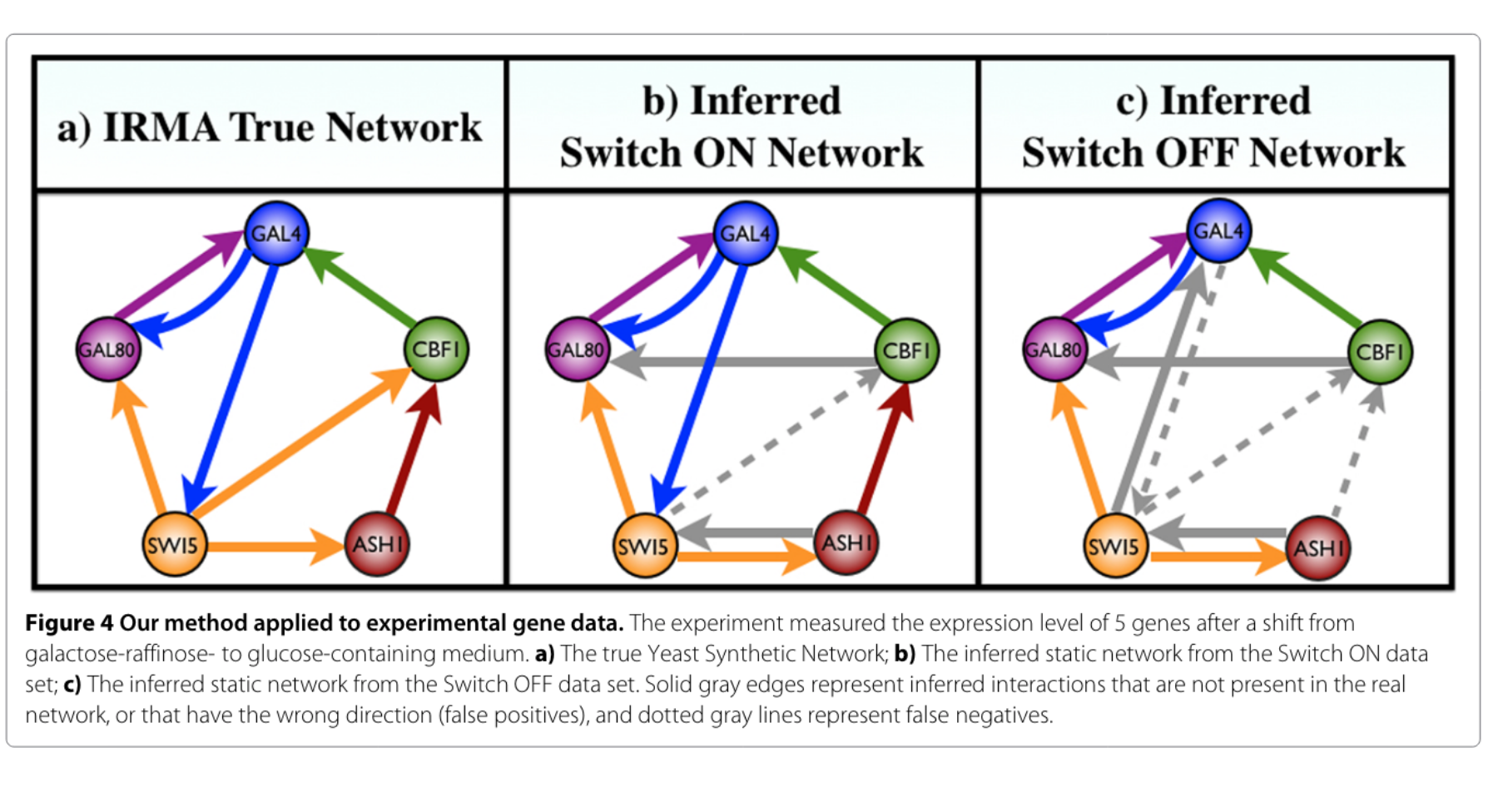
In this paper we developed a novel reverse-engineering method for inferring gene regulatory networks within the Boolean modeling framework. This method is innovative in that the inference problem is framed in computational algebra terms to enable precise algebraic characterization of the models search space.
Paola Vera-Licona, Abdul Jarrah, Luis D Garcia-Puente, John McGee, Reinhard Laubenbacher
BMC Systems Biology volume 8, Article number 37 (2014)
This paper was Selected as Editors Picks
List of publications
NETISCE: A Network-Based Tool for Cell Fate Reprogramming
Lauren Marazzi, Milan Shah, Shreedula Balakrishnan, Ananya Patil, Paola Vera-Licona
npj Systems Biology and Applications volume 8, Article number 21 (2022)
Macrophage mediated recognition and clearance of Borrelia burgdorferi elicits MyD88-dependent and -independent phagosomal signals that contribute to phagocytosis and inflammation
Sarah J. Benjamin, Kelly L. Hawley, Paola Vera-Licona, Carson J. La Vake, Jorge L. Cervantes, Yijun Ruan, Justin D. Radolf and Juan C. Salazar
BMC Immunology volume 22, Article number 32 (2021)
OCSANA+ Optimal Control and Simulation of Signaling Networks from Network Analysis
Lauren Marazzi, Andrew Gainer-Dewar, Paola Vera-Licona
Bioinformatics, Volume 36, Issue 19 (2020)
Identification of a nerve-associated, lung-resident interstitial macrophage subset with distinct localization and immunoregulatory properties
Basak B Ural,Stephen T Yeung, Payal Damani-Yokota, Joseph C Devlin, Maren de Vries, Paola Vera-Licona, Tasleem Samji, Catherine M Sawai, Geunhyo Jang, Oriana A Perez, Quynh Pham, Leigh Maher Png Loke, Meike Dittmann, Boris Reizis and Kamal M. Khanna
Science Immunology, Vol 5, Issue 45 (2020)
Benchmarking time-series data discretization on inference methods
Yuezhe Li, Tiffany Jann, Paola Vera-Licona
Bioinformatics, Volume 35, Issue 17 (2019)
Chromatin interaction networks revealed unique connectivity patterns of broad H3K4me3 domains and super enhancers in 3D chromatin
Asa Thibodeau, Eladio J Márquez, Dong-Guk Shin, Paola Vera-Licona, Duygu Ucar
Scientific Reports volume 7, Article number 14466 (2017)
CD169+ macrophages orchestrate innate immune responses by regulating bacterial localization in the spleen
Oriana Perez, Stephen Yeung, Paola Vera-Licona, Pablo Romagnoli, Tasleem Samji, Basak B Ural, Leigh Maher, Masato Tanaka, Kamal M Khanna
Science Immunology, Vol 2, Issue 16 (2017)
The minimal hitting set generation problem: algorithms and computation
Andrew Gainer-Dewar, Paola Vera-Licona
SIAM Journal on Discrete Mathematics Vol. 31 (2017)
QuIN: a web server for querying and visualizing chromatin interaction networks
Asa Thibodeau, Eladio J Márquez, Oscar Luo, Yijun Ruan, Francesca Menghi, Dong-Guk Shin, Michael L Stitzel, Paola Vera-Licona, Duygu Ucar
PLoS Comput Biol 12, 6 (2016)
AlgoRun: a Docker-based packaging system for platform-agnostic implemented algorithms
Abdelrahman Hosny, Paola Vera-Licona, Reinhard Laubenbacher, Thibauld Favre
Bioinformatics, Volume 32, Issue 15 (2016)
An Algebra-Based Method for Inferring Gene Regulatory Networks
Paola Vera-Licona, Abdul Jarrah, Luis D Garcia-Puente, John McGee, Reinhard Laubenbacher
BMC Systems Biology volume 8, Article number 37 (2014)