Software

NETISCE is a computational tool for identifying cell fate reprogramming targets in static networks. In combination with machine learning algorithms, NETISCE estimates the attractor landscape and predicts reprogramming targets using Signal Flow Analysis and Feedback Vertex Set Control, respectively.
npj Systems Biology and Applications volume 8, Article number 21 (2022)
The installation, tutorials, information for installing the Galaxy Project version of NETISCE, and walkthroughs for reproducing our analysis results are available at http://veraliconalab.org/Netisce/
The NETISCE Nextflow pipeline version, the Docker image documentation, and data are available Github Repository

OCSANA+ is a Cytoscape app for identifying nodes to drive the system toward a desired long-term behavior, prioritizing combinations of interventions in large-scale complex networks, and estimating the effects of node perturbations in signaling networks, all based on the analysis of the network’s structure.
Bioinformatics, Volume 36, Issue 19 (2020)
The installation, tutorials, and a walkthrough example for reproducing our analysis results are available at Website http://veraliconalab.org/OCSANA-Plus/
Access OCSANA+ from the Cytoscape App store. Access the source code in our Github Repository

DiscreeTest is a two-step evaluation metric for ranking discretization methods for time-series data.
Bioinformatics, Vol. 35, Issue 17 (2019)
Source code and data sets and computations to reproduce our analysis can be found in our Github Repository

Software implementation of several algorithms to find inclusion-minimal hitting sets (MHSs) for a given family of sets.
SIAM Journal on Discrete Mathematics Vol. 31 (2017)
The source code of a collection of implemented algorithms for solving the MHS generation problem, easy-to-use AlgoRun containers for these algorithms, and data sets and computations to reproduce our analysis can be found in our Github Repository
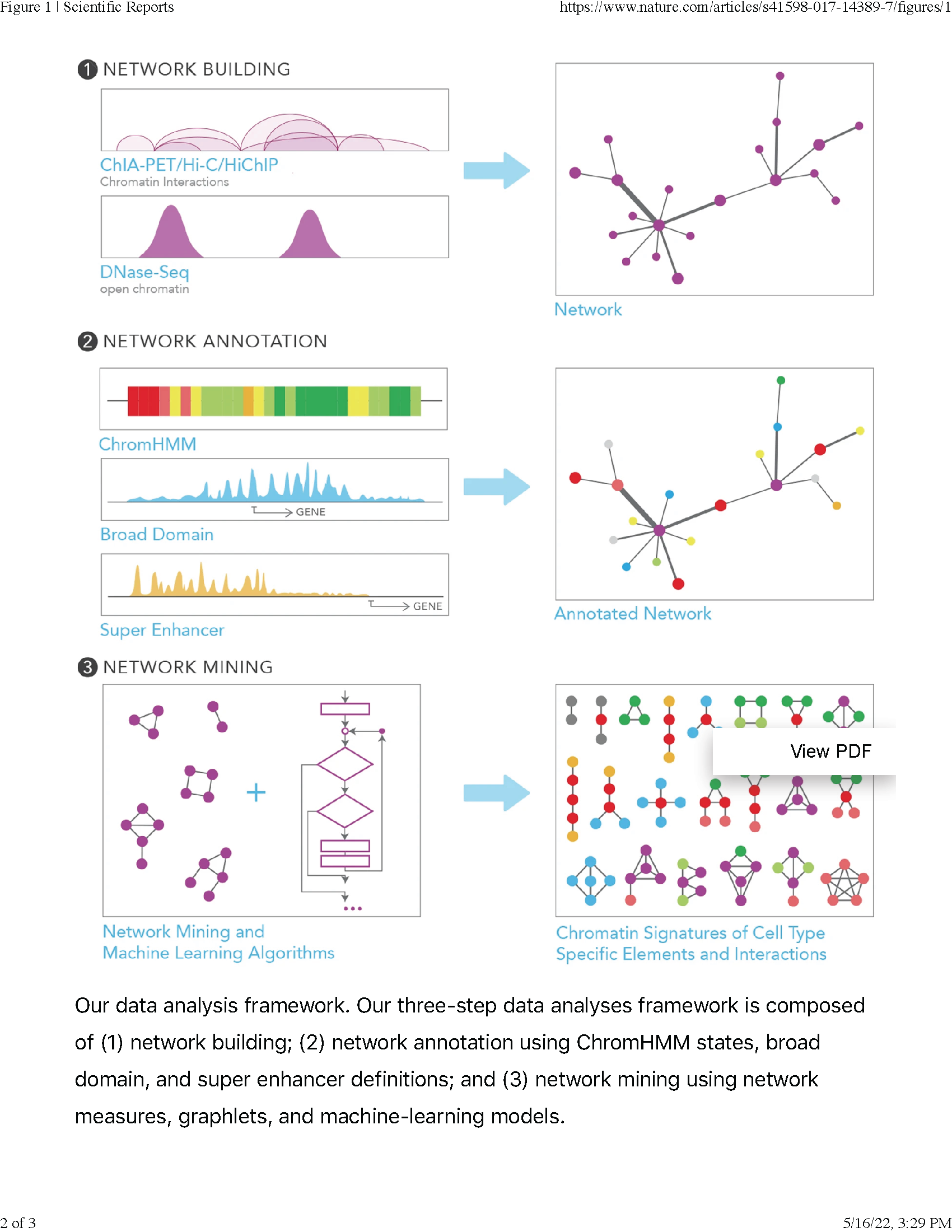
This tool was created in collaboration with the Ucar Lab. QuIN is a tool for querying and visualizing chromatin interaction networks. This is the first publicly available software for analysis of chromatin interaction networks that is web-accessible and easy to use, making it suitable for molecular biologists with no programming experience.
Source code is available at Ucar Lab Github Repository

AlgoRun is a useful Docker-based software tool for the scientific community developing software in their research endeavors that enables an easy way to prepare software tools with a user-friendly execution, cross-platform portability, and simple distribution. This tool was developed in collaboration with the Laubenbacher Lab and Thibauld Favre.
Bioinformatics, Volume 32, Issue 15 (2016)
The AlgoRun website with documentation and a collection of algorithms can be accessed at http://algorun.org/
Source code is available in the Github Repository

A reverse-engineering method for inferring gene regulatory networks within the Boolean modeling framework. This method is innovative in that the inference problem is framed in computational algebra terms to enable precise algebraic characterization of the models search space.
BMC Systems Biology volume 8, Article number 37 (2014)
Source code is available in the Github Repository

OCSANA is a software designed to identify and prioritize optimal and minimal combinations of intreventions to disrupt the paths between source nodes and target nodes. The tool is part of BiNoM 2.8. OCSANA was developed while in the Systems Biology of Cancer Lab at Institut Curie.
Bioinformatics, Volume 29, Issue 12, 15 (2013)
The installation, tutorials, and a walkthrough example for reproducing our analysis results are available at the website http://bioinfo-out.curie.fr/projects/ocsana/OCSANA.html
Source code is available in the Github Repository